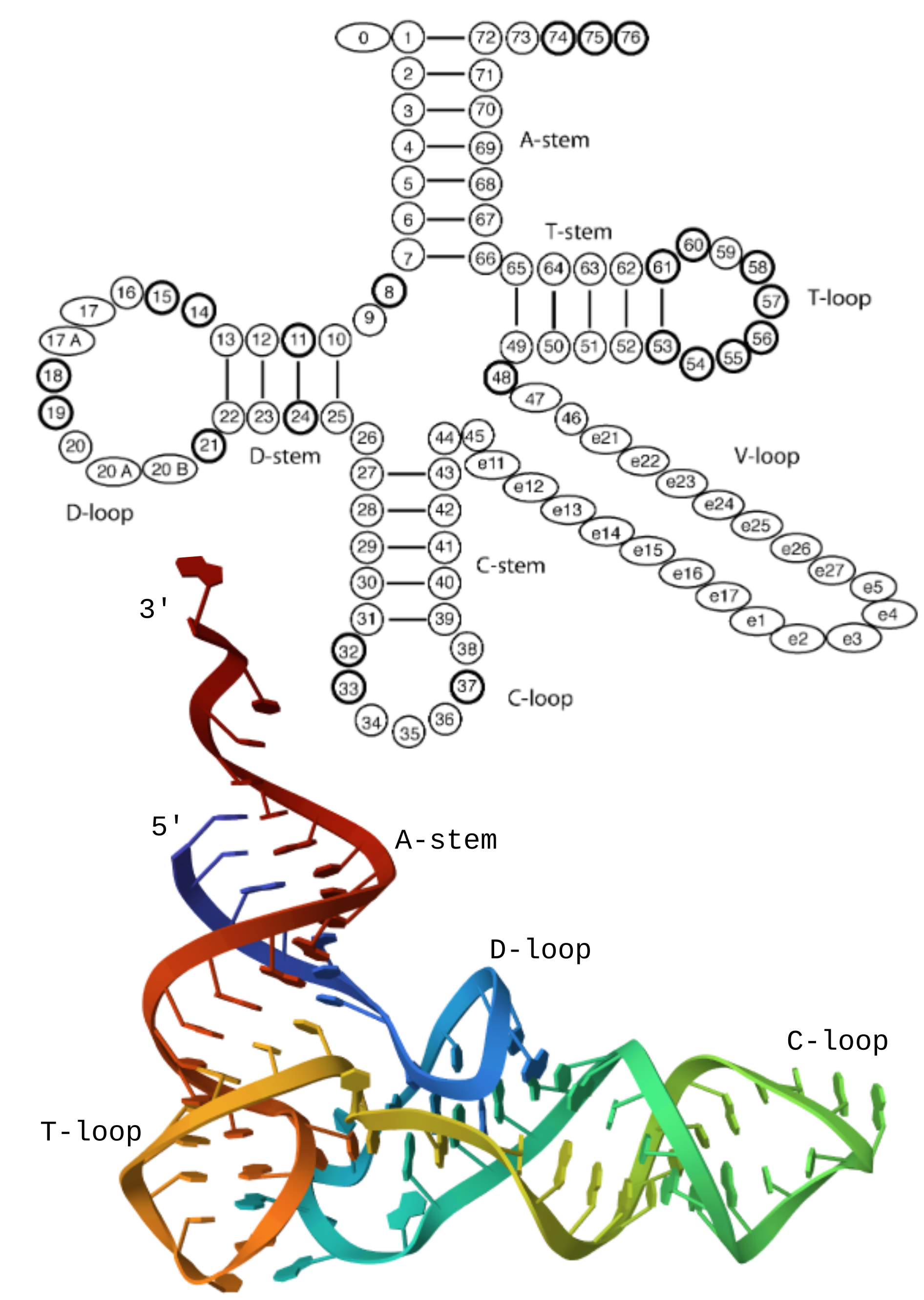
 tRNA molecular structure. The top diagram shows the components of a tRNA and
the canonical numbering (from the Laslett2004 paper, Figure 1). The colored
image below shows the crystal structure of yeast phenylalanine tRNA
(1EHZ). The 5’ end is blue and transforms to red at the 3’. I made this
image on the PDB website.
tRNA molecular structure. The top diagram shows the components of a tRNA and
the canonical numbering (from the Laslett2004 paper, Figure 1). The colored
image below shows the crystal structure of yeast phenylalanine tRNA
(1EHZ). The 5’ end is blue and transforms to red at the 3’. I made this
image on the PDB website.
One of the objectives of morloc is to replace the file-oriented paradigm of
bioinformatics with a simpler functional approach. I have long advocated for
rewriting existing bioinformatics software in this way. In the morloc paper,
currently under review, I present a case study where I developed an influenza
clade classification pipeline from scratch, allowing me to tailor the design to
the morloc framework. While this demonstrated the power of morloc, it did
not address the practical challenges of transitioning existing codebases. To
meet this need, I have chosen to refactor the ARAGORN tRNA prediction tool
(Laslett 2004). This tool is a good candidate because it is popular, with nearly
3,000 citations, it is complex enough to be challenging, and it is functionally
simple taking sequences as input and producing tRNAs as output.
ARAGORN scans DNA sequence for encoded transfer RNAs (tRNAs) and transfer-messenger RNAs (tmRNAs). These molecules play central roles in life. tRNAs ferry amino acids to the growing polypeptide chain as mRNA is translated to protein. tmRNAs help rescue stalled ribosomes and tag incomplete proteins for degradation. These RNAs are among the most ancient of all recognizable structures in biology. They likely evolved with the first livings things before ribosomes, DNA and proteins (Macé 2016, Guyomar 2019). In humans today, mutations in tRNAs can cause many diseases (Abbot 2014). Thus accurately identifying them is important in molecular evolutionary studies, basic biological research, genetic medicine.
tRNAs share a core “clover-leaf” structure (see figure on right). The 5’ and 3’ ends of the RNA are joined in the A-stem. This is where the amino acid attaches in tRNAs. The highly-conserved T and D loops preserve structure. The variable loop (V-loop) ranges in length from 3 to 26 bases (according to ARAGORN’s permissive parameters) and can stabilize the RNA structure and tune inter-molecular interactions (e.g. binding to the ribosome). In tRNAs, the C-loop contains the anticodon that recognizes 3-letter patterns in protein templates and translates them into amino acids. Mitochondrial tRNAs (mtRNAs)
tmRNAs deviate from the canonical “clover-leaf”. The anticodon region is replaced with a long mRNA-like strand and they lack the hairpin structure where the D-loop would be.
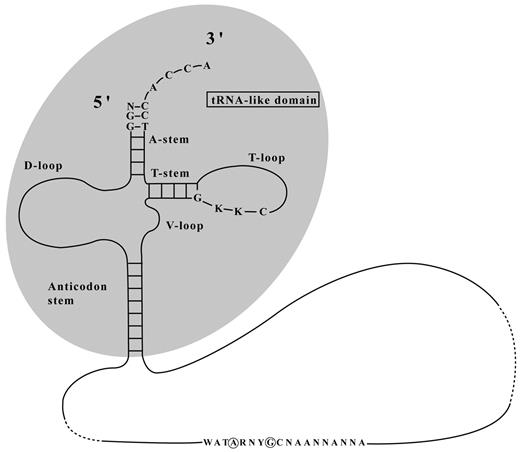 tmRNAs have a long mRNA-like loop in place of an anticodon triplet and lack
a D-loop. Image from (Laslett 2004).
tmRNAs have a long mRNA-like loop in place of an anticodon triplet and lack
a D-loop. Image from (Laslett 2004).
Mitochondrial tRNA are wonderfully deviant cousins of genomic tRNAs. They break motifs conserved for billions of years and may loose entire T or D loops. Few of them passed as tRNAs in the 2004 version of ARAGORN. In 2008, however, a new version of ARAGORN, released with the aptly named package ARWEN (Laslett 2008), added over 3000 lines of code designed to lock onto these patterns.
This tutorial series explores the transformation of the ARAGORN application into a modular and flexible library. The goal is to enhance its usability in larger systems and improve its interchangeability with similar tools.
In Part 1, we’ll examine ARAGORN’s current input/output structure and propose a simplified approach. The aim is to streamline the tool to accept a single DNA string as input and produce a list of tRNA records as output, leaving format choices and processing details to the user.
Part 2 will focus on ARAGORN’s parameters. We’ll discuss how to separate various functionalities from the core package, such as output type selection, input processing, and result filtering. These features will be pruned from the core library and delegated to more general purpose libraries.
Part 3 will cover the reorganization of ARAGORN’s codebase. We’ll explore how
the original 12,000-line application was reduced to a 1,000-line library. The
resulting code can be compiled into a shared library that can serve as a
foundation for larger applications both with and without morloc.
Part 4 will demonstrate the integration of new ARAGORN library into the morloc
ecosystem. We’ll create a morloc module that imports the ARAGORN shared
library and then expand it into a larger trna module. This section will
showcase the ease of building libraries in morloc and creating modular
bioinformatics tools.
While I will be nitpicking issues with ARAGORN in this series, I do consider it to be one of the best bioinformatics tools. It is fast and stable, compiles easily, has decent usage statements, flexible IO options, and a focused scientific objective. As a command line tool, it is excellent. My objective is to show how a better tool can be created with less work and composed more easily with other tools. My fight is with the file-oriented paradigm and ARAGORN is a clean example from that circle.
References
Laslett, Dean, and Bjorn Canback. “ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences.” Nucleic acids research 32.1 (2004): 11-16.
Macé, Kevin, and Reynald Gillet. “Origins of tmRNA: the missing link in the birth of protein synthesis?.” Nucleic Acids Research 44.17 (2016): 8041-8051.
Guyomar, Charlotte, and Reynald Gillet. “When transfer‐messenger RNA scars reveal its ancient origins.” Annals of the New York Academy of Sciences 1447.1 (2019): 80-87.
Abbott, Jamie A., Christopher S. Francklyn, and Susan M. Robey-Bond. “Transfer RNA and human disease.” Frontiers in genetics 5 (2014): 158.